Identification and Speciation of Aspergillus in Clinical and Environmental Samples using Culture-Based Methods: A Laboratory-based Study in Hyderabad
Abstract
Background: Aspergillosis is increasingly recognized as a serious opportunistic infection, particularly among immunocompromised patients. Accurate species-level identification of Aspergillus is critical for selecting appropriate antifungal therapy and implementing effective infection control measures. This study aimed to identify and characterize Aspergillus species isolated from both clinical specimens and indoor air samples at a tertiary care hospital in Hyderabad, Telangana, India. Methods: Over a one-year period, a total of 709 specimens—including nasal mucosa swabs, tissue biopsies (lung, FESS), blood samples, nail clippings, and passive indoor air settle plates—were processed in the microbiology laboratory. Direct microscopic examination was performed using KOH mount, periodic acid–Schiff stain, and calcofluor white stain. Specimens showing filamentous fungi were cultured on Sabouraud dextrose agar, potato dextrose agar, corn meal agar, Czapek medium, and malt extract agar, and incubated at 25–30 °C for 7–10 days. Species identification was based on detailed assessment of colony morphology and microscopic features (conidiophore structure, vesicle shape, phialide arrangement, and conidial ornamentation), following CLSI guidelines. Results: Out of 709 specimens, Aspergillus species were isolated from 239 samples (33.7%). Thirteen species were identified, with A. flavus being the most common (33.4%), followed by A. niger (26.7%), A. fumigatus (19.2%), A. nidulans (4.6%), A. glaucus (3.3%), A. terreus (2.9%), A. versicolor (2.1%), A. calidoustus (1.7%), A. glabrata (1.7%), A. parasiticus (1.2%), A. clavatus (1.3%), A. ochraceus (0.8%), and A. tanneri (0.8%). A. flavus predominated among both clinical and environmental isolates. Conclusion: Conventional morphological methods combined with the use of multiple culture media proved effective for species-level identification of Aspergillus. Routine surveillance of Aspergillus species in both clinical and environmental samples can guide targeted antifungal therapy and support proactive infection control in healthcare environments.
KEYWORDS: Aspergillus, Speciation, Differential media, Clinical specimens, Environmental surveillance
INTRODUCTION
The genus Aspergillus now comprises of 339 recognized mould species, widely distributed in soil, decaying vegetation, and ambient air. Of these, approximately 40 species are known to cause human disease, ranging from superficial infections such as otomycosis and cutaneous lesions to more severe manifestations like chronic necrotizing and invasive pulmonary aspergillosis, particularly among immunocompromised individuals. 1, 2 The most commonly isolated species in clinical practice include A. fumigatus, A. niger, A. terreus, and A. flavus. 3
Aspergillus fumigatus remains the leading cause of invasive aspergillosis worldwide. This is attributed to its small conidial size, which facilitates deep penetration into the respiratory tract, and its thermotolerance. 4, 5 In contrast, A. flavus is the predominant species in hot, arid regions such as the Indian subcontinent, owing to its ability to survive at higher temperatures and its distinct virulence factors, including potent mycotoxin production and strong allergenic potential. 6, 7
A. terreus also exhibits inherent resistance to amphotericin B 8 , which highlights the critical need for accurate species-level identification to guide appropriate antifungal therapy.
Species differentiation in routine microbiology laboratories primarily relies on the assessment of colony morphology including growth rate, surface and reverse pigmentation, and texture as well as microscopic features such as conidial ornamentation and the arrangement of vesicles and phialides. These methods are standardized under the Clinical and Laboratory Standards Institute (CLSI) guidelines (M38-A2). 9
Balajee et al. demonstrated that careful macroscopic and microscopic examination of isolates grown on Sabouraud dextrose agar, Czapek’s solution agar, and malt extract agar enables accurate differentiation of common pathogenic species such as A. fumigatus, A. niger, A. flavus, and A. terreus. 3
While molecular techniques such as polymerase chain reaction (PCR) assays targeting species-specific DNA sequences 10 and matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS) for proteomic profiling offer rapid and highly precise identification 11 , their implementation requires specialized instrumentation, molecular infrastructure, and skilled personnel, limiting their accessibility in many resource-limited settings. 12 As a result, conventional culture-based methods combined with microscopic evaluation remain vital tools for Aspergillus speciation in clinical mycology laboratories.
Environmental surveillance of Aspergillus within healthcare facilities is equally important to infection control, given that airborne spores can cause nosocomial outbreaks, particularly during periods of hospital construction or ventilation system failures. Gravity-settle plates and volumetric air sampling methods have demonstrated a strong correlation between elevated airborne fungal counts and the incidence of invasive aspergillosis in high-risk units. 13 Recent reviews emphasize the importance of routine environmental monitoring and the implementation of strict control measures to reduce patient exposure to pathogenic Aspergillus species. 14
The present laboratory-based study was undertaken to identify the species distribution of Aspergillus isolates recovered from routine clinical specimens (including nasal mucosa swabs, tissue biopsies, blood samples, and nail clippings) and passive indoor air samples at a tertiary care hospital in Hyderabad, India.
MATERIALS AND METHODS
Study design and setting: This laboratory-based cross-sectional study was conducted in the Microbiology Laboratory of Osmania General Hospital, Hyderabad, Telangana, India, between 1 April 2023 and 31 March 2024. The study aimed to determine the prevalence and species distribution of Aspergillus in routinely processed clinical and environmental specimens over a defined one-year period.
Specimen selection: All clinical (nasal mucosa swabs, tissue obtained during functional endoscopic sinus surgery [FESS], lung biopsies, blood, and nail clippings) and environmental (indoor air) specimens submitted for mycological evaluation during the study period were included. No additional sampling was performed outside routine diagnostic submissions.
Air sampling: Passive air sampling was conducted monthly using the settle-plate method in high-risk wards and shared hospital areas. Sterile 90 mm Petri dishes containing Sabouraud dextrose agar (SDA; HiMedia, Mumbai, India) were exposed at a height of 1.5 meters for 30 minutes. Plates were then sealed and processed along with clinical specimens.
Direct microscopy: non-tissue specimens were examined using 10% potassium hydroxide (KOH) wet mounts. Tissue biopsies were assessed using periodic acid–Schiff (PAS) staining and calcofluor white (CFW) fluorescence microscopy to detect filamentous fungal elements.
Culture and incubation: Specimens positive for filamentous fungi on microscopy were inoculated in parallel onto SDA, Potato dextrose agar (PDA), Czapek’s solution agar (CZA), and Malt extract agar (MEA), all procured from HiMedia (Mumbai, India). Cultures were incubated at 25 ± 2 °C for up to 10 days, Observations like colony diameter, surface and reverse pigmentation, texture and growth rate—were recorded on Days 3, 5 and 7.
Phenotypic identification: Slide cultures were prepared on CZA and stained with lactophenol cotton blue (LPCB). Microscopic identification was based on conidiophore morphology, vesicle configuration, phialide arrangement, and conidial ornamentation, examined under 400× magnification. Species determination followed CLSI M38-A2 guidelines and standard taxonomic references.
Quality control: A. fumigatus ATCC 204305 was included as an internal control to verify culture media performance, incubation parameters, and morphological identification consistency.
Data analysis: Frequencies and species proportions were described in number and percentages and calculated using MS Excel 2021.
RESULTS
A total of 709 specimens were processed over the one-year study period; Aspergillus was isolated from 239 samples (33.7%), comprising of 149 (62.3%) clinical specimens and 90 (37.7%) environmental (passive indoor air) samples. Thirteen Aspergillus species were identified, with A. flavus, A. niger, and A. fumigatus collectively accounting for 79.3% of all isolates (Table 1).
|
Species |
Frequency |
Percentage (%) |
|
A. flavus |
80 |
33.5 |
|
A. niger |
64 |
26.8 |
|
A. fumigatus |
46 |
19.2 |
|
A. nidulans |
11 |
4.6 |
|
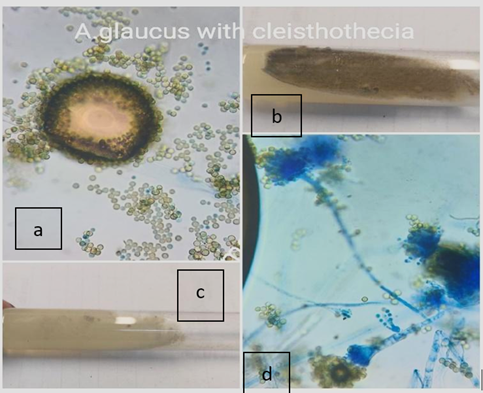
A. glaucus |
8 |
3.3 |
|
A. terreus |
7 |
2.9 |
|
A. versicolor |
5 |
2.1 |
|
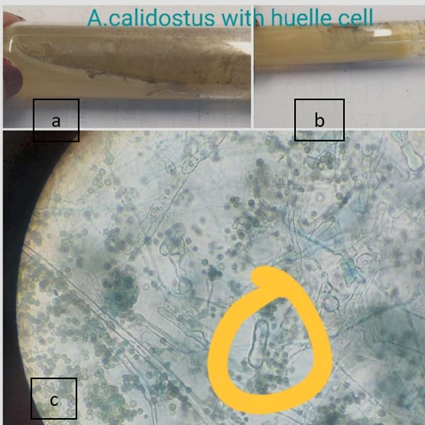
A. calidoustus |
4 |
1.7 |
|
A. glabrata |
4 |
1.7 |
|
A. parasiticus |
3 |
1.2 |
|
A. clavatus |
3 |
1.3 |
|
A. ochraceus |
2 |
0.8 |
|
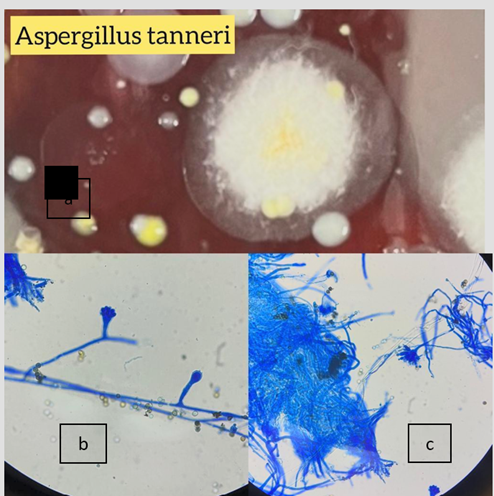
A. tanneri |
2 |
0.8 |
Macroscopic and microscopic observations
Colonies grown on Sabouraud dextrose agar (SDA), Czapek’s solution agar (CZA), and malt extract agar (MEA) exhibited distinct species-specific features such as pigmentation, texture, and growth rate. Microscopic examination revealed characteristic structures like conidiophore branching patterns, vesicle shapes, phialide arrangements, and conidial ornamentation, which were in accordance with CLSI M38-A2 guidelines. Characteristic features of the three most frequently isolated species are summarized in Table 2.
|
Species |
Medium |
Obverse colour |
Reverse colour |
Key microscopic features |
|
A. flavus |
SDA |
Yellow-green |
Olive-green |
Rough, echinulate conidia; biseriate phialides |
|
CZA |
Yellow-green |
Hyaline |
||
|
MEA |
Dark green |
Hyaline |
||
|
A. fumigatus |
SDA |
Smoky-grey |
Green |
Smooth, globose conidia; uniseriate phialides |
|
CZA |
Grey |
Hyaline |
||
|
MEA |
Blue-grey |
Hyaline |
||
|
A. terreus |
SDA |
Beige |
Cinnamon-buff |
Cylindrical, rough-walled conidia; columnar uniseriate phialides |
|
CZA |
Yellow-orange |
Light yellow |
||
|
MEA |
Light yellow |
Yellowish-brown |
Species distribution by specimen type
The species distribution across the seven specimen categories is shown in Table 3. A. flavus was the most frequently isolated species from both clinical specimens (47/149; 31.5%) and environmental air samples (33/90; 36.7%), followed by A. niger in both categories.
|
Species |
Blood |
Nasal mucosa |
FESS tissue |
Middle meatus |
Lung biopsy |
Nail clippings |
Air samples |
Total |
|
A. flavus |
1 |
17 |
18 |
7 |
2 |
2 |
33 |
80 |
|
A. niger |
0 |
12 |
10 |
11 |
0 |
3 |
28 |
64 |
|
A. fumigatus |
1 |
7 |
14 |
8 |
5 |
0 |
11 |
46 |
|
A. nidulans |
0 |
3 |
1 |
0 |
6 |
0 |
1 |
11 |
|
A. glaucus |
0 |
1 |
1 |
1 |
2 |
0 |
3 |
8 |
|
A. terreus |
0 |
2 |
1 |
0 |
3 |
0 |
1 |
7 |
|
A. versicolor |
0 |
0 |
1 |
1 |
2 |
1 |
1 |
5 |
|
A. calidoustus |
0 |
0 |
0 |
0 |
1 |
0 |
3 |
4 |
|
A. glabrata |
0 |
0 |
2 |
0 |
0 |
2 |
0 |
4 |
|
A. parasiticus |
0 |
0 |
0 |
0 |
0 |
0 |
3 |
3 |
|
A. clavatus |
0 |
0 |
1 |
0 |
0 |
0 |
2 |
3 |
|
A. ochraceus |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
2 |
|
A. tanneri |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
2 |
|
Total |
2 |
42 |
47 |
30 |
21 |
8 |
83 |
239 |
In clinical specimens (n = 149): nasal and FESS samples together accounted for 89/149 (59.7%) of clinical isolates whereas in environmental samples (n = 90): the predominant isolates were A. flavus (36.7%) and A. niger (31.1%).
The most common species isolated from nasal mucosa was A. flavus (n=17) followed by A. niger (n=12). FESS samples showed A. flavus (n=18) followed by A. fumigatus (n=14) and A. niger (n=10). Middle meatus showed presence was A. niger (n=11) followed by A. fumigatus (n=8) and A. flavus (n=7). Majority of the species from air samples belonged to A. flavus (n=33) followed by A. niger (n=28) and A. fumigatus (n=11).
DISCUSSION
In this laboratory-based cross-sectional study, Aspergillus species were isolated from 33.7% (239/709) of routinely processed clinical and environmental specimens. While direct comparison with large surveillance datasets is limited by differing denominators and inclusion criteria, previous Indian studies report broadly similar species distributions. Bansal et al. 15 documented A. flavus (40%), A. fumigatus (35%), and A. niger (25%) as predominant isolates in 200 culture-positive clinical samples. Similarly, Chowdhary et al. 16 , in a referral chest hospital in Delhi, reported Aspergillus isolation in 25.7% of clinical specimens, with A. flavus (45.4%), A. fumigatus (32.4%), and A. terreus (6.6%) being most frequent. Additionally, this finding is consistent with research conducted by Diba K. et al. 17 and Balajee SA et al. 3
Our finding that A. flavus (33.5%) was the most frequently isolated species emphasizes its epidemiological predominance in the tropical Indian context. This may be due to its inherent thermotolerance, environmental resilience, and the production of mycotoxins and allergens that enhance virulence. 6
Environmental surveillance using passive air sampling recovered Aspergillus from 90 samples (37.7% of total isolates), with A. flavus (36.7%) and A. niger (31.1%) being most common. These results are in consistence with a settle-plate surveillance study from a hospital in Puducherry, where A. niger (25.0%), A. flavus (17.3%), and A. fumigatus (13.5%) were the dominant airborne fungi. 18 Discrepancies in species prevalence may reflect differences in microenvironmental variables such as air circulation, humidity, temperature, and human activity levels. This highlights the importance of standardized environmental surveillance to guide infection-control strategies. 19
Species-level identification is critical for clinical relevance. Although A. terreus accounted for only 2.9% of isolates in our study, its intrinsic resistance to amphotericin B 8 highlights the risk of therapeutic failure when misidentified. Additionally, the emergence of azole-resistant A. fumigatus, potentially driven by agricultural fungicide exposure, is of growing concern. A recent Indian study reported a 1.7% prevalence of triazole resistance in clinical isolates 16 , stressing the value of routine antifungal susceptibility testing for both clinical and environmental strains.
This study has several limitations. Species identification was based exclusively on morphological criteria without molecular confirmation, which may limit taxonomic precision. Antifungal susceptibility profiling was not performed, excluding assessment of local resistance patterns. Furthermore, passive settle-plate sampling may underestimate airborne spore loads compared to active volumetric methods. Future studies should incorporate advanced diagnostic modalities such as polymerase chain reaction (PCR), matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS), and antifungal susceptibility testing to strengthen epidemiological insights and therapeutic guidance.
In summary, our findings are consistent with the predominance of A. flavus in both clinical and environmental settings in India. Despite resource constraints, conventional morphology-based methods remain effective for routine speciation in diagnostic laboratories. However, integrating molecular tools and resistance surveillance into routine practice is essential to enhance patient outcomes and reinforce infection prevention efforts.
CONCLUSION
This laboratory-based cross-sectional study demonstrated that Aspergillus species were isolated from one-third of routine clinical and environmental specimens, with A. flavus, A. niger, and A. fumigatus collectively accounting for nearly 80% of all isolates. The predominance of A. flavus in both patient-derived and indoor air samples highlights its epidemiological significance in tropical climates and suggests a potential role in healthcare-associated transmission.
Conventional morphological identification using differential culture media remains a practical and cost-effective method for species-level identification, particularly in resource-constrained settings. However, to improve patient management and strengthen infection control, future initiatives should incorporate active volumetric air sampling, molecular diagnostic techniques (e.g., PCR, MALDI-TOF MS), and routine antifungal susceptibility testing. These measures will enable more precise surveillance, facilitate early detection of antifungal resistance, and guide targeted antifungal therapy.
Disclosure
Funding: None
Conflict of Interest: None Declared
Author Contribution: All the authors involved in study have contributed equally at all stages of work
Acknowledgements: I would like to express my appreciation to all those who have supported and contributed to the completion of this project


